Programmatic Data Access Guide
All data from the NASA National Snow and Ice Data Center Distributed Active Archive Center (NSIDC DAAC) is directly accessible through our HTTPS file system using Wget or curl. This article provides basic command line instructions for accessing data using this method.
Additionally, a large selection of NSIDC DAAC data is available through our Application Programming Interface (API). The API enables you to request data with specific temporal and spatial filters, as well as subset, reformat, and reproject select data sets. This article also includes API guidance and a link to a Jupyter Notebook that helps you construct and download your data request.
Regardless of how users access data from the NSIDC DAAC, an Earthdata Login account is required. Please visit the Earthdata Login registration page to register for an account before getting started.
For questions about programmatic access to NSIDC DAAC data, please contact NSIDC User Services at nsidc@nsidc.org.
HTTPS File System URLs
To access data with a browser or Wget, you’ll need the HTTPS File System URL. NSIDC DAAC is migrating its data collections to NASA’s Earthdata Cloud through 2026. As data moves to the cloud, HTTPS access for the cloud archive will be available via the HTTPS File System link on each data set’s landing page. Users can still access the legacy HTTPS access locations until they’re retired. While both cloud and legacy HTTPS locations are available, the legacy HTTPS access option can be accessed using the URLs listed below.
The top-level URLs for access through the Legacy HTTPS File System are:
https://n5eil01u.ecs.nsidc.org
https://daacdata.apps.nsidc.org
Specific URLs for commonly accessed NASA data collections moving to Earthdata Cloud are:
AMSR Unified and AMSR-E: https://n5eil01u.ecs.nsidc.org/AMSA/
High Mountain Asia (HMA): https://n5eil01u.ecs.nsidc.org/HMA/
ICESat-2: https://n5eil01u.ecs.nsidc.org/ATLAS/
ICESat/GLAS: https://n5eil01u.ecs.nsidc.org/GLAS/
MEaSUREs: https://n5eil01u.ecs.nsidc.org/MEASURES/
MODIS-Aqua: https://n5eil01u.ecs.nsidc.org/MOSA/
MODIS-Terra: https://n5eil01u.ecs.nsidc.org/MOST/
NASA SnowEx: https://n5eil01u.ecs.nsidc.org/SNOWEX/
Operation IceBridge/LVIS: https://n5eil01u.ecs.nsidc.org/ICEBRIDGE/
SMAP: https://n5eil01u.ecs.nsidc.org/SMAP/
SMMR-SSM/I-SSMIS: https://n5eil01u.ecs.nsidc.org/PM/
VIIRS: https://n5eil01u.ecs.nsidc.org/VIIRS/
As legacy HTTPS access locations are retired, the URLs above will stop working and the links will be removed. For the latest updates on data collection migration, please see https://nsidc.org/data/earthdata-cloud/news.
HTTPS access: Wget instructions for Mac and Linux
Step 1: Set up authentication
Store your Earthdata Login credentials (username <uid> and password <password>) for authentication in a .netrc file in your home directory.
Example commands showing how to set up a .netrc file for authentication:
echo 'machine urs.earthdata.nasa.gov login <uid> password <password>' >> ~/.netrcchmod 0600 ~/.netrcReplace <uid> and <password> with your Earthdata Login username and password (omit brackets). Windows bash users should also replace the forward slash ( / ) with a backslash ( \ ) in the above commands.
Alternatively, skip Step 1 by using these options in the Wget command in Step 2:--http-user=<uid> --http-password=<password>
or--http-user=<uid> --ask-password
Step 2: Download data
Use a Wget command to download your data. The URL prefix depends on the data set: either https://n5eil01u.ecs.nsidc.org or https://daacdata.apps.nsidc.org. Check the data set’s landing page and select the ‘HTTPS File System’ card to find the correct URL. Here are two examples for each URL type:
Example 1: Wget command to download SMAP L4 Global Daily 9 km EASE-Grid Carbon Net Ecosystem Exchange, Version 6 data for 07 October 2019:
wget --load-cookies ~/.urs_cookies --save-cookies ~/.urs_cookies --keep-session-cookies --no-check-certificate --auth-no-challenge=on -r --reject "index.html*" -np -e robots=off https://n5eil01u.ecs.nsidc.org/SMAP/SPL4CMDL.006/2019.10.07/SMAP_L4_C_mdl_20191007T000000_Vv6042_001.h5Example 2: Wget command to download Canadian Meteorological Center (CMC) Daily Snow Depth Analysis Data, Version 1 data for the year 2020:
wget --load-cookies ~/.urs_cookies --save-cookies ~/.urs_cookies --keep-session-cookies --no-check-certificate --auth-no-challenge=on -r --reject "index.html*" -np -e robots=off https://daacdata.apps.nsidc.org/pub/DATASETS/nsidc0447_CMC_snow_depth_v01/Snow_Depth/Snow_Depth_Daily_Values/GeoTIFF/cmc_sdepth_dly_2020_v01.2.tif
Files download to a directory named after the HTTPS host, "n5eil01u.ecs.nsidc.org" or "daacdata.apps.nsidc.org". You can modify how the files are stored by specifying the following Wget options:
-nd (or --no-directories)
-nH (or --no-host-directories)
The GNU Wget 1.20 Manual provides more details on Wget options.
HTTPS access: Wget instructions for Windows
The following instructions apply to Wget executed through Windows Command Prompt. For Windows bash, please see the above Mac and Linux instructions.
Step 1: Create a cookie file
Create a text file ("mycookies.txt") to store the website cookies returned from the HTTPS server. Store this in the wget installation directory.
Step 2: Download data
Use a Wget command to download your data. In the following example, replace <uid> and <password> with your Earthdata Login username and password (omit brackets).
The URL prefix depends on the data set: either https://n5eil01u.ecs.nsidc.org or https://daacdata.apps.nsidc.org. To determine which to use, visit the data set’s landing page, select the 'HTTPS File System' card, and use the URL shown at the top. Here are two examples for each URL type:
Example 1: Wget command to download SMAP L4 Global Daily 9 km EASE-Grid Carbon Net Ecosystem Exchange, Version 6 data for 07 October 2019:
wget --http-user=<uid> --http-password=<password> --load-cookies mycookies.txt --save-cookies mycookies.txt --keep-session-cookies --no-check-certificate --auth-no-challenge -r --reject "index.html*" -np -e robots=off https://n5eil01u.ecs.nsidc.org/SMAP/SPL4CMDL.006/2019.10.07/SMAP_L4_C_mdl_20191007T000000_Vv6042_001.h5Example 2: Wget command to download Canadian Meteorological Center (CMC) Daily Snow Depth Analysis Data, Version 1 data for the year 2020:
wget --http-user=<uid> --http-password=<password> --load-cookies mycookies.txt --save-cookies mycookies.txt --keep-session-cookies --no-check-certificate --auth-no-challenge -r --reject "index.html*" -np -e robots=off https://daacdata.apps.nsidc.org/pub/DATASETS/nsidc0447_CMC_snow_depth_v01/Snow_Depth/Snow_Depth_Daily_Values/GeoTIFF/cmc_sdepth_dly_2020_v01.2.tifFiles download to a directory named after the HTTPS host, "n5eil01u.ecs.nsidc.org" or "daacdata.apps.nsidc.org". You can modify how the files are stored by specifying the following Wget options:
-nd (or --no-directories)
-nH (or --no-host-directories)
The GNU Wget 1.20 Manual provides more details on Wget options.
HTTPS access: cURL instructions
Step 1: Set up authentication
Follow the Step 1 Wget instructions for Mac and Linux above. Windows users should replace the forward slashes ( / ) with backslashes ( \ ) in the same commands.
Step 2: Download data
Use a curl command to download your data.
The URL prefix depends on the data set: either https://n5eil01u.ecs.nsidc.org or https://daacdata.apps.nsidc.org. To determine which to use, visit the data set’s landing page, select the 'HTTPS File System' card, and use the URL shown at the top. Here are two examples for each URL type:
Example 1: curl command to download a single file of SMAP L4 Global Daily 9 km EASE-Grid Carbon Net Ecosystem Exchange, Version 6 data for 07 October 2019:
curl -b ~/.urs_cookies -c ~/.urs_cookies -L -n -O https://n5eil01u.ecs.nsidc.org/SMAP/SPL4CMDL.006/2019.10.07/SMAP_L4_C_mdl_20191007T000000_Vv6042_001.h5Example 2: Wget command to download Canadian Meteorological Center (CMC) Daily Snow Depth Analysis Data, Version 1 data for the year 2020:
curl -b ~/.urs_cookies -c ~/.urs_cookies -L -n -O https://daacdata.apps.nsidc.org/pub/DATASETS/nsidc0447_CMC_snow_depth_v01/Snow_Depth/Snow_Depth_Daily_Values/GeoTIFF/cmc_sdepth_dly_2020_v01.2.tifThe -O option in the curl command downloads the file to your current working directory.
NSIDC DAAC's data access and service API
Note: While a large selection of NSIDC DAAC data can be accessed using the API (e.g., AMSR-E/AMSR2, ICESat-2/ICESat, MODIS, SMAP, VIIRS) not all NSIDC DAAC data can be accessed this way.
The API provided by the NSIDC DAAC enables programmatic data access using specific temporal and spatial filters. It offers the same subsetting, reformatting, and reprojection services available for select data sets through NASA Earthdata Search. These options can be requested in a single access command without the need to script against our data directory structure. This API is beneficial for those who frequently download NSIDC DAAC data and want to incorporate data access commands into their analysis or visualization code. If you’re new to the API, we recommend exploring this service using our Jupyter Notebook. Further guidance on API format and usage examples are provided below.
Data Access Jupyter notebook
The Jupyter notebook available on Github allows you to explore the data coverage, size, and customization services available for your data sets. It generates API commands usable from a command line, browser, or within the Notebook itself. No Python experience is necessary—each code cell prompts you through configuring your data request. The README offers multiple options for running the notebook, including Binder, Conda, and Docker. The Binder button in the README allows you to easily explore and run the notebook in a shared cloud computing environment without the need to install dependencies on your local machine. For bulk data downloads, we recommend running the notebook locally via Conda or Docker, or using the print API command option if running via Binder.
Data customization services
All data sets can be accessed and filtered based on time and area of interest. We also offer customization services (i.e. subsetting, reformatting, and reprojection) for certain NSIDC DAAC collections. Please visit the following pages for detailed service availabilty:
How to format the API endpoint
Programmatic API requests are formatted as HTTPS URLs that contain key-value-pairs that specify the service operations:
https://n5eil02u.ecs.nsidc.org/egi/request?<key-value-pairs separated with &>
There are three categories of API key-value-pairs, as described below. Please reference the lookup Table of Key-Value-Pair (KVP) Operands for Subsetting, Reformatting, and Reprojection Services to see all keyword options available for use.
Search keywords
These keywords include the data set short name (e.g. MOD10A1), version number, time range of interest, and area of interest used to filter* the file results.
Customize keywords
These keywords include subsetting by variable, time, and spatial area*, along with reformatting and reprojection services. These subsetting services modify the data outputs by limiting the data based on the values specified.
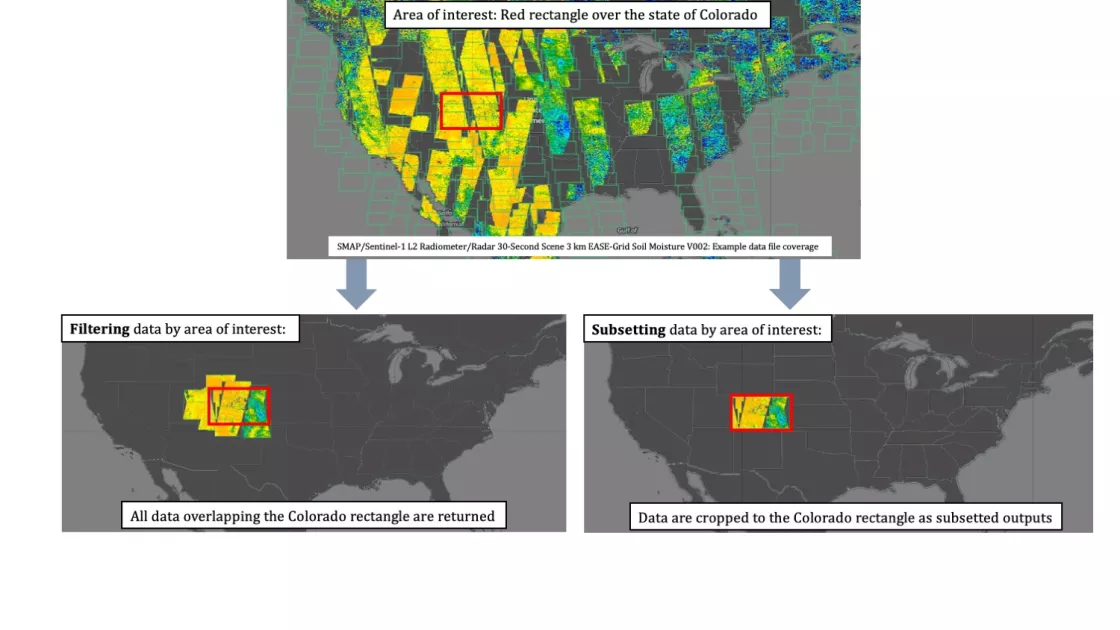
*See Figure 1 for more information on the difference between spatial filtering and spatial subsetting.
Configuration keywords
These keywords include data delivery method (either asynchronous or synchronous), email address, and the number of files returned per API endpoint. The asynchronous data delivery option allows concurrent requests to be queued and processed without the need for a continuous connection; orders will be delivered to the specified email address. Synchronous requests automatically download the data as soon as processing is complete; this is the default if no method is specified. The file limits differ between the two options:
Maximum files per synchronous request = 100
Maximum files per asynchronous request = 2000
If the number of files is not specified, the API will only deliver 10 files by default. You can adjust the number of files returned using the page_size and page_num keywords if you are experiencing performance issues.
To request data by rectangular area of interest, use the bounding_box keyword to specify the search filter bounds. If you also want to subset the data to that area of interest, include the bbox subsetting keyword with coordinates identical to the bounding_box search filter keyword.
Figure 1. Data filtering requests return whole files based on overlapping coverage, as seen on the lower left-hand map. Data subsetting requests crop the data files to the area of interest, resulting in subsetted file outputs, as seen on the lower right-hand map. All data can be spatially filtered using the API, whereas only select data sets offer spatial subsetting. Spatial subsetting requests require the spatial filtering keyword to avoid errors.
API endpoint examples
Downloading data in its native format (no customization)
To request data without subsetting, reformatting, or reprojection services (using only spatial and/or temporal filtering), you must use the agent=NO parameter, as demonstrated in the following examples:
Example 1: AMSR-E/AMSR2 Unified L2B Global Swath Ocean Products, Version 1
- Temporal extent: 2021-12-07 12:00:00 to 2021-12-07 15:00:00
- Maximum file number specified (page_size)
- No customization services (agent=NO)
- Result: A zip file returned synchronously with 5 science files and associated metadata
Example 2: MODIS/Terra Snow Cover Daily L3 Global 500m SIN Grid, Version 61
- Temporal extent: 2018-05-31 17:03:36 to 2018-06-01 06:47:53
- Spatial extent: Iceland
- Maximum file number specified
- Native data requested (no customization services)
- Result: A zip file returned synchronously containing 3 science files and associated metadata
Example 3: MEaSUREs Greenland Ice Velocity: Selected Glacier Site Velocity Maps from InSAR, Version 4
- Temporal extent: 01 January 2017 to 31 December 2018
- Spatial extent: Western Greenland coast
- No customization services
- No metadata requested
- Note: Because 133 files will be returned, two endpoints are needed to capture all results (100 files in the first request, 33 in the second)
https://n5eil02u.ecs.nsidc.org/egi/request?short_name=NSIDC-0481&version=4&time=2017-01-01T00:00:00,2018-12-31T00:00:00&bounding_box=-52.5,68.5,-47.5,69.5&agent=NO&INCLUDE_META=N&page_size=100&page_num=1
https://n5eil02u.ecs.nsidc.org/egi/request?short_name=NSIDC-0481&version=4&time=2017-01-01T00:00:00,2018-12-31T00:00:00&bounding_box=-52.5,68.5,-47.5,69.5&agent=NO&INCLUDE_META=N&page_size=100&page_num=2
Downloading customized data (reformatting, spatial and parameter subsetting, and reprojection)
Example 4: SMAP L3 Radiometer Global Daily 36 km EASE-Grid Soil Moisture, Version 9
- Temporal extent: 2008-06-06 to 2018-06-07
- Spatial extent: Colorado
- Customizations: GeoTIFF reformatting, spatial subsetting, parameter subsetting, and geographic reprojection
- Maximum file number specified
- Results: A zip file returned synchronously with 2 reformatted, subsetted, and reprojected files
Example 5: MODIS/Terra Snow Cover Monthly L3 Global 0.05Deg CMG, Version 61
- Temporal extent: 2015-01-01 to 2015-10-01
- Customizations: GeoTIFF reformatting and parameter subsetting
- Maximum file number specified
- Results: A zip file returned synchronously with 10 reformatted and subsetted files
Using the API curl
Example 1: MODIS example 5 above:
curl -b ~/.urs_cookies -c ~/.urs_cookies -L -n -O -J --dump-header response-header.txt "https://n5eil02u.ecs.nsidc.org/egi/request?short_name=MOD10CM&version=6&format=GeoTIFF&time=2015-01-01,2015-10-01&Coverage=/MOD_CMG_Snow_5km/Snow_Cover_Monthly_CMG"Example 2: ATLAS/ICESat-2 L3A Land Ice Height, Version 2, with variable subsetting for all data collected on 01 August 2019 over Pine Island Glacier using an uploaded Shapefile* of the glacier boundary:
curl -b ~/.urs_cookies -c ~/.urs_cookies -L -n -O -J -F "shapefile=@glims_download_PIG.zip" "https://n5eil02u.ecs.nsidc.org/egi/request?short_name=ATL06&version=002&time=2019-08-01,2019-08-02&polygon=-86.69021127827942,-74.83792495569011,-90.06285424412475,-73.99476421422878,-94.2786579514314,-73.71371063374167,-96.52708659532829,-74.13529100447232,-100.04025635141716,-73.99476421422878,-102.28868499531404,-74.556871375203,-101.30499746360915,-74.83792495569011,-101.86710462458338,-75.11897853617722,-103.97500647823671,-75.54055890690788,-101.1644706733656,-76.38371964836921,-101.1644706733656,-76.94582680934343,-96.66761338557184,-77.50793397031765,-96.38655980508473,-77.64846076056124,-97.37024733678962,-78.07004113129187,-95.40287227337984,-78.49162150202255,-95.40287227337984,-79.33478224348386,-92.73286325875229,-80.03741619470165,-88.9386399221763,-79.05372866299675,-91.88970251729097,-78.35109471177898,-90.76548819534253,-77.78898755080476,-92.31128288802165,-77.36740718007411,-91.04654177582964,-77.086353599587,-91.74917572704742,-76.80530001909989,-87.3928452294972,-75.96213927763856,-86.83073806852298,-75.68108569715145,-86.69021127827942,-74.83792495569011&coverage=/gt1l/land_ice_segments/h_li,/gt1l/land_ice_segments/latitude,/gt1l/land_ice_segments/longitude,/gt1r/land_ice_segments/h_li,/gt1r/land_ice_segments/latitude,/gt1r/land_ice_segments/longitude,/gt2l/land_ice_segments/h_li,/gt2l/land_ice_segments/latitude,/gt2l/land_ice_segments/longitude,/gt2r/land_ice_segments/h_li,/gt2r/land_ice_segments/latitude,/gt2r/land_ice_segments/longitude,/gt3l/land_ice_segments/h_li,/gt3l/land_ice_segments/latitude,/gt3l/land_ice_segments/longitude,/gt3r/land_ice_segments/h_li,/gt3r/land_ice_segments/latitude,/gt3r/land_ice_segments/longitude,/quality_assessment"*This example includes uploading a zipped Shapefile to be used for subsetting, in addition to filtering by simplified polygon coordinates based on the uploaded file. This functionality is currently only available for ICESat-2 data. This Shapefile is available through the NSIDC Global Land Ice Measurements from Space (GLIMS) database. Direct download access available here.
Example 3: Large file requests
The following bash script example demonstrates a request using the asynchronous delivery method. This method is managed by our ordering system and doesn't require a continuous connection to execute. We recommend this method for requests that require long processing times or exceed the 100-file synchronous delivery method limit. This script requests the same MEaSUREs data in example 3 above. It retrieves the order number and order status, and delivers the data programmatically in addition to an email notification.
#!/bin/bash
#make the request
curl -s -b ~/.urs_cookies -c ~/.urs_cookies -L -n -o 'request.xml' 'https://n5eil02u.ecs.nsidc.org/egi/request?request_mode=async&short_name=NSIDC-0481&version=1&time=2017-01-01T00:00:00,2018-12-31T00:00:00&bounding_box=-52.5,68.5,-47.5,69.5&agent=NO&INCLUDE_META=N&page_size=2000&email=yes'
#parse the response for requestid.
REQUESTID=`grep orderId request.xml | awk -F '<|>' '{print $3}'`
#poll for status
for i in {1..360}
do
curl -s -b ~/.urs_cookies -c ~/.urs_cookies -L -n -o 'response.xml' 'https://n5eil02u.ecs.nsidc.org/egi/request/'"$REQUESTID"
RESPONSESTATE=`grep status response.xml | awk -F '<|>' '{print $3}'`
if [ "$RESPONSESTATE" != "processing" ]
then
break
fi
sleep 10;
done
#get the zips
for i in `grep downloadUrl response.xml | grep -v '\.html<' | awk -F '<|>' '{print $3}'`; do
curl -s -b ~/.urs_cookies -c ~/.urs_cookies -L -n -O "$i"
done
Last updated: November 2024